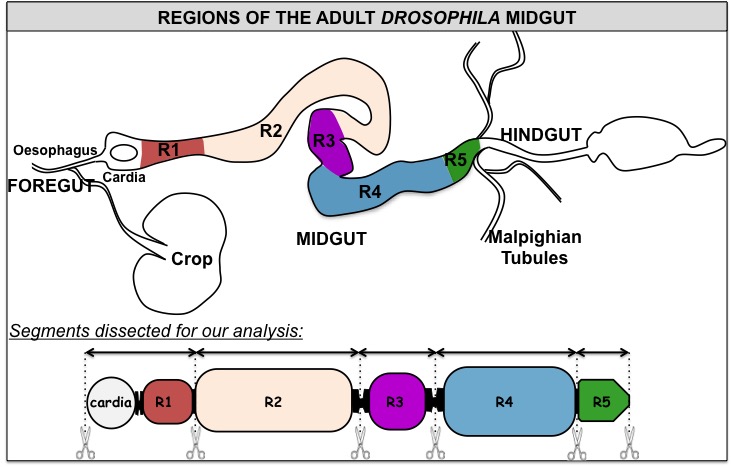
How were the midgut regions were separated?
The midguts were manually dissected under a microscope and regions were separated by hand based on the anatomical features of the main midgut regions identified in Buchon et al 2013, Cell reports. More information on gut regions can be found at flygut.epfl.ch
What does switched on or off mean?
In the infection data, we present fold induction of guts in infected conditions compared to unchallenged conditions. However if the gene was not originally detected in this cell type, but is now expressed, we refer to it as switched on. If the gene is expressed in basal conditions but no more detected upon infection, we refer to it as switched off. Also, when genes are regulated more than 1000 times, we refer to it as switched on or off, in order to still see milder regulation in different cell types.
RPKM (Reads per kilobase per million)
RPKM values are a measure of transcript abundance and therefore gene expression. They represent the number of reads obtained by RNAseq for a specific gene normalized both by gene length and by the total number of reads per sample.The formula to calculate RPKM (reads mapping to the genome per kilobase of transcript per million reads sequenced) :
RPKM = 10^9 x C/NL RPKM=(number of mapping reads)*1000bp*1 million reads/[(length of transcript)*(number of total reads )].
10^9 corresponds to 1000bp transcript * 1 million reads.
Why mention gene rpkm / RpL32 rpkm?
This ratio is indicative of the RT-qPCR ratio you could measure with RpL32 (Rp49) as a reference gene.